The Computational Biomedical Imaging Group (CBIG) pursues research on the development of new algorithms and mathematical tools for the reconstruction and post-processing of medical and biological images. Topics of interest include image reconstruction, post-processing and quantification in the context of a range of modalities including magnetic resonance imaging, near-infrared spectroscopic imaging and electron microscopy.Research efforts are taking place at two complementary levels (a)fundamental and mathematical aspects of imaging (b) application-oriented projects in collaboration with researchers in medicine and biology. Some of the current projects are listed below.
Accelerated cardiac MRI
|
The acquisition of cardiac perfusion MRI with high spatio-temporal resolution and spatial coverage is challenging. We propose a novel scheme to significantly accelerate the acquisitions in cardiac perfusion MRI. The proposed scheme exploits the low-rank and sparsity of the data to accelerate cardiac perfusion MRI by a factor of 8 or more. The top image shows the reconstructions from ten fold accelerated data, while the bottom image is the original cardiac perfusion time series. Note that the ten fold accelerated data accurately captures the temporal dynamics without significant distortions. See our paper1
and paper2
for more information.
|

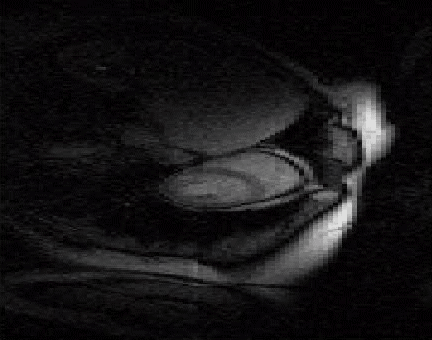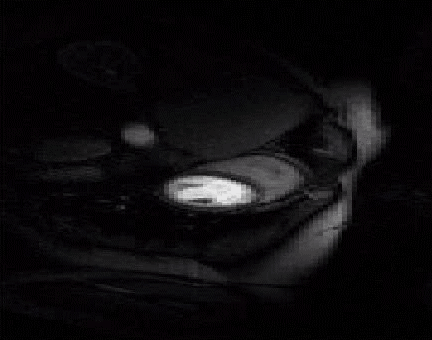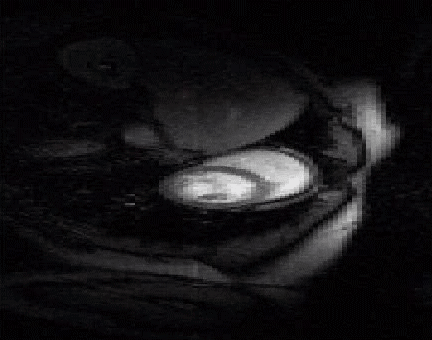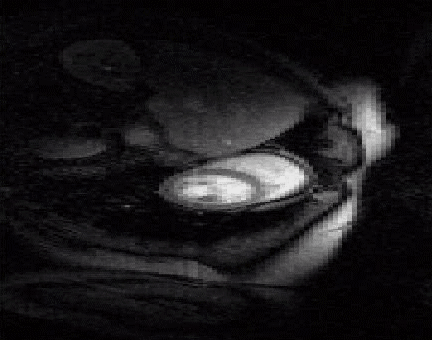
|
Improved reconstruction of MR spectroscopic imaging data
|
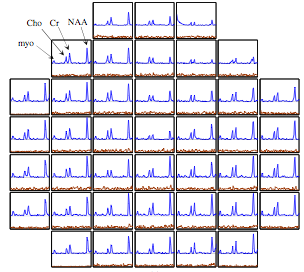
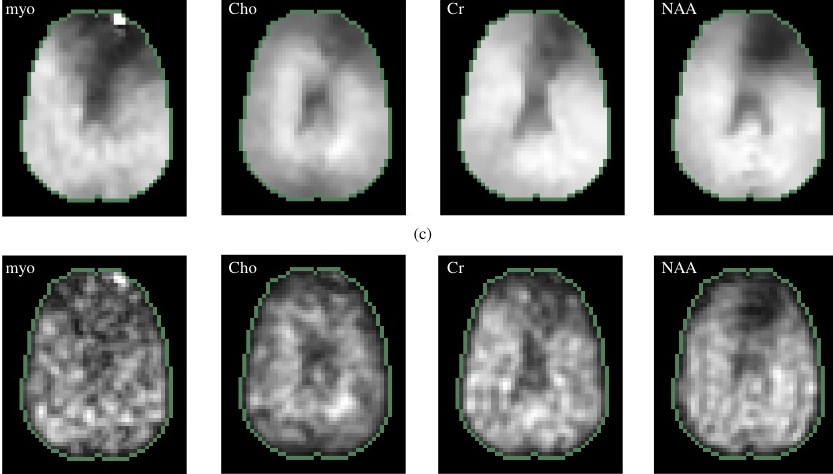
Magnetic resonance spectroscopic imaging (MRSI) is emerging
as a promising tool to determine microscopic tumor spread in glioma, which can be capitalized to target focal therapies and hence improve treatment outcome. Current MRSI schemes are often corrupted by a range of artifacts due to inhomogeneity of the main magnetic field and unsuppressed fat & water. We propose a novel regularized reconstruction scheme to exploit high resolution spatial priors and sparse spectral priors to overcome these challenges. The top figure shows the spectra reconstructed using the proposed scheme. The second row indicates the metabolite estimates derived using the proposed scheme from a glioma patient, while the bottom row is estimated using the state of the art method. See paper1
and paper2
for more information.
|
|
Algebraic decomposition of fat and water
|
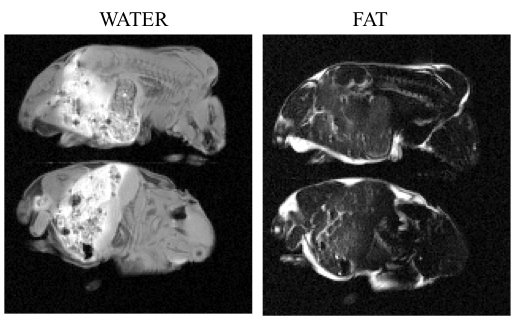
The decomposition of MRI data to generate water and fat images has several applications in medical imaging, including fat suppression and quantification of visceral fat. We introduce a novel algorithm to overcome the problems associated with current analytical and iterative decomposition schemes such as lack of flexibility and convergence to local minima. See our paper for more information.See our paper for more information. We are currently applying this assay to study the alterred lipid metabolism in obesity. Shown in left are the water and fat images of obese rat pups (21 days old).
|
|
|
Reconstruction of functional activations from NIRSI data
|
|
|
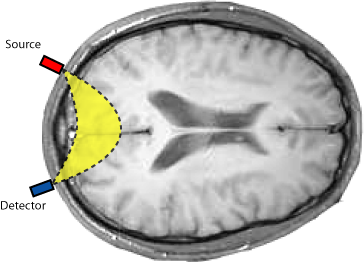
Near infrared spectroscopic imaging offers remarkable biochemical specificity over other functional imaging schemes such as functional MRI. However the attainable spatial resolution with this technique is rather limited. We introduced a new algorithm for the reconstruction of functional brain activations from near infrared spectroscopic imaging (NIRSI) data. See our paper for more information.See our paper
for more information.
|
|
Reconstruction of DNA filaments from cryo-electron micrographs
|
|
|
Cryo-electron microscopy is a technique used to image biomolecules like DNA. The main challenge in reconstructing the shape of the molecule is the low signal to noise ratio and the low number of projections. We introduce a 3-D parametric active contour algorithm for the shape estimation of DNA molecules from stereo cryo-electron micrographs. See our paper1 and paper2 for more information.
|

|

|
Efficient algorithms for parametric snakes
|
|
|
Parametric active contour models are one of the preferred approaches for image segmentation because of their computational efficiency and simplicity. However, they have a few drawbacks which limit their performance. We identified these problems and proposed efficient workarounds, thus making them even more powerful. A JAVA implementation of this algorithm is available here. See our paper for more information.
|

|
Design of steerable feature detectors
|
|
|
Feature detection is a key element of many image processing and computer vision applications. We propose a general approach to derive optimal steerable feature detectors of arbitrary order. This approach gives better orientation selective detectors and hence result in better detections. A JAVA implementation of this algorithm as well as online demo is available here. See our paper for more information.
|
 |
 |
|
|

